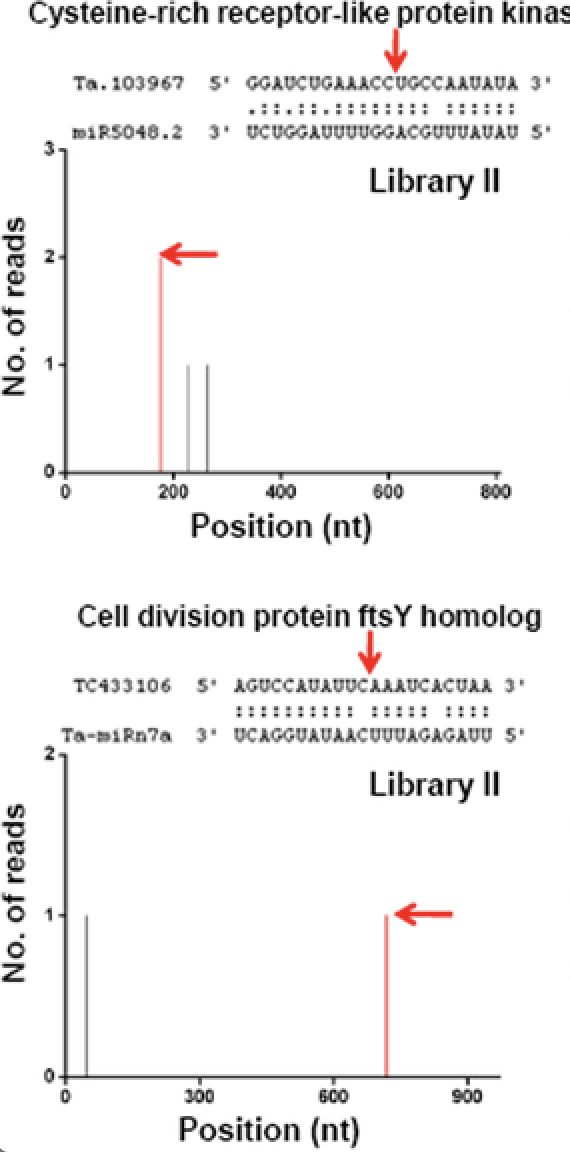
Degradome sis a modified version of 5'-equencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing method using as Illumina's SBS technology. Degradome sequencing provides a comprehensive means of analyzing patterns of RNA degradation.
Degradome sequencing has been used to identify: microRNA (miRNA) cleavage sites, because miRNAs can cause endonucleolytic cleavage of mRNA by extensive and often perfect complementarity to mRNAs. Degradome sequencing reveals many known and miRNA (siRNA) targets nd miRNA-dervied cleavages.
Sequencing using HiSEQ 2500
Single end 50 base Read length
40 Million Paired-end Reads
Standard Bioinformatics Included